Principal Investigator:
Jacob Geri, Assistant Professor of Pharmacology
Background & Unmet Need
- Measuring binding affinities between ligands and proteins is crucial for understanding the potency, selectivity, and specific effects of molecules in complex biological systems
- Existing methods for measuring binding affinity are typically accomplished in vitro, requiring purified proteins and prior knowledge of the interaction, limiting use in complex samples like live cells or serum
- Proximity labeling technologies can identify novel interactions in vivo (e.g., protein-protein, protein-DNA, protein-RNA) but lack quantitative binding affinity measurements
- Unmet Need: Robust, proteome-wide, and quantitative measurement of binding affinities in complex biological samples such as live cells, cell lysates, and organ extracts
Technology Overview
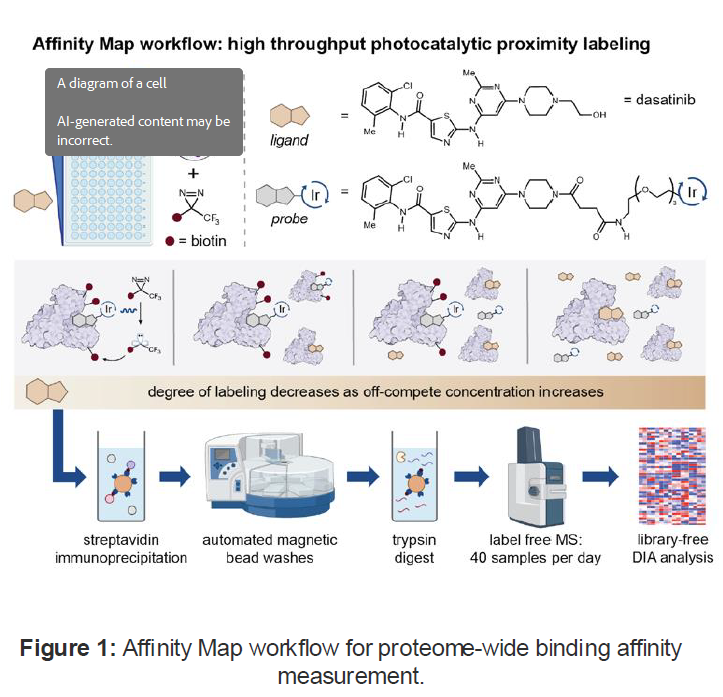
- The Technology: Affinity Map is a novel platform that enables robust measurement of binding affinities between ligands and proteins across the proteome using photocatalytic labeling and high-throughput mass spectrometry
- A “hot” ligand conjugated to a photocatalyst and a “cold” unmodified ligand are used to covalently label proteins, enabling competitive binding assays that yield quantitative affinity measurements
- Affinity Map is applicable to major classes of ligands: small molecules, linear and cyclic peptides, proteins
- PoC Data: Affinity Map measured Kd values for the protein interactions of dasatinib, a tyrosine inhibitor, and JQ1, a selective bromodomain binding compound, with high accuracy and selectivity, even identifying previously unknown interactions
- Affinity Map was used to measure Kd values for the macrocyclic cRBDfk peptide and human IgG on live cell surfaces, finding close concurrence with reported literature values
Technology Applications
- Global small molecule-protein, peptide-protein, or protein-protein binding affinity determination
- Can be leveraged in complex biological systems such as cell lysates, protein extracts, and live cells
- Use in early drug discovery to screen novel chemical matter or profile interactions of small molecule candidates, including binding affinity, target engagement, and proteome-wide off-target binding affinities
Technology Advantages
- Measures binding affinities across all protein classes, including membrane proteins and non-kinase targets
- Maintains a low false detection negative rate, outperforming methods like CETSA and limited proteolysis
- Provides quantitative binding affinities using competitive kinetics, offering insights beyond qualitative fold enrichment data from other platforms
Resources
Intellectual Property
Patents
- Provisional application filed
Cornell Reference
- 11190
Contact Information

For additional information please contact
Jamie Brisbois
Manager, Business Development and Licensing
Phone: (646) 921-4743
Email: jamie.brisbois@cornell.edu

