Principal Investigator:
Lishomwa (Lish) Ndhlovu, Herbert J. and Ann L. Siegel Distinguished Professor of Medicine
Scott Bowler, Senior Bioinformatician
Background & Unmet Need
- Immune checkpoint blockade (ICB) therapies, such as anti-PD-1 and anti-CTLA antibodies, have demonstrated great success in a variety of cancers
- Despite eligibility for immune checkpoint blockade (ICB) therapy increasing over 30x, response rates continue to barely exceed 12%
- Biomarkers for predicting patient responsiveness currently in use, including PD-L1 expression, tumor mutational burden, and microsatellite instability fail consistently classify patients with high accuracy
- A variety of host factors influencing ICB efficacy have been proposed, including human endogenous retroviruses (HERVs), a type of transposable element contributing to oncogenesis
- Unmet Need: Additional biomarkers that enable more accurately prediction of patient responsiveness to ICB therapies
Technology Overview
- The Technology: HERV-K/HML2 provirus expression as a biomarker for predicting response to ICB therapy
- The Discovery: An extreme boosted gradient machine learning model (XGBOOST) was developed to identify bacterial and viral signatures associated with ICB response among RNA from 139 melanoma, NSCLC, RCC, and GBM tumor samples
- Support vector machine framework and recursive feature elimination were used to develop a separate model (SVM) based on DNA from 385 melanoma, NSCLC, and RCC tumor samples were used to train had available TME-derived DNA
- PoC Data: The models identified identified HERV-K/HML2 expression as a key factor for ICB response
- The XGBoost model identified 8 proviruses which classify participants by ICB response (AUC-ROC=0.801)
Technology Applications
- Method to identify patients who are likely to respond to ICB therapy in melanoma, NSCLC, RCC, and/or GBM
- Use as a biomarker for patient selection or stratification for ICB clinical trials
Technology Advantages
- Broad applicability across multiple solid tumor types
- HERV-K expression may provide an independent biomarker to complement existing biomarkers for ICB therapy, such as PD-L1 expression, microsatellite instability, and tumor mutational burden
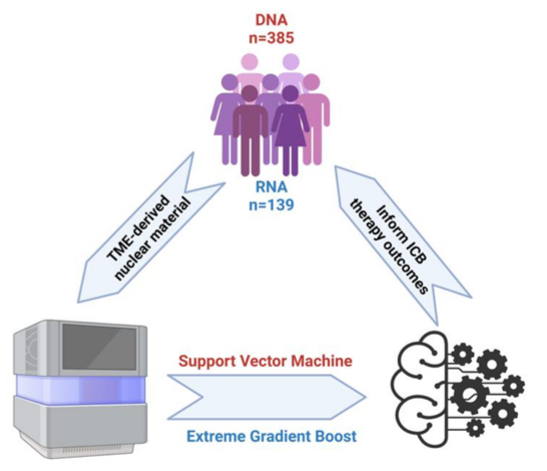
Figure: Graphical abstract of XGBOOST and SVM model development
Intellectual Property
Patents
- Provisional Application Filed
Cornell Reference
- 11459
Contact Information

For additional information please contact
Jamie Brisbois
Manager, Business Development and Licensing
Phone: (646) 921-4743
Email: jamie.brisbois@cornell.edu

